Gallery
You are here :
Some pictures issued from docking studies and protein-ligand interactions, giving some ideas (not exhaustive, not chronological) of our production. The related bibliographic references are shown at the bottom of the page.

Ferutinin docked in two conformations of ERa nuclear receptor
Docking poses of Ferutinin in ERa binding cavities. Ferutinin (FRT, green) docking poses are shown in 1G50 (A, chain A, ERa, brown, closed cavity) and 1XPC (B, chain A, ERa, purple, opened cavity) structures, including the corresponding co-crystallized ligands EST (gold, agonist) and AIT (purple, antagonist). The clipping planes of aligned structures are the same, and set using lateral chains of GLU 353 and ARG394 (left), PHE 404 is also shown (left-up).
Docking poses of Ferutinin in ERa binding cavities. Ferutinin (FRT, green) docking poses are shown in 1G50 (A, chain A, ERa, brown, closed cavity) and 1XPC (B, chain A, ERa, purple, opened cavity) structures, including the corresponding co-crystallized ligands EST (gold, agonist) and AIT (purple, antagonist). The clipping planes of aligned structures are the same, and set using lateral chains of GLU 353 and ARG394 (left), PHE 404 is also shown (left-up).
Susbtrate (THT, 1BVR) and inhibitor (GEQ, 1P44) superposition after protein alignment
Superimposition of GEQ (blue) and substrate analog (purple) molecules in the InhA binding site (PDB IDs respectively 1P44 and 1BVR). Coloring according the kd Hydrophobicity scale for each amino acid from dodger blue to the most hydrophilic, to white, to orange red for the most hydrophobic.
Superimposition of GEQ (blue) and substrate analog (purple) molecules in the InhA binding site (PDB IDs respectively 1P44 and 1BVR). Coloring according the kd Hydrophobicity scale for each amino acid from dodger blue to the most hydrophilic, to white, to orange red for the most hydrophobic.
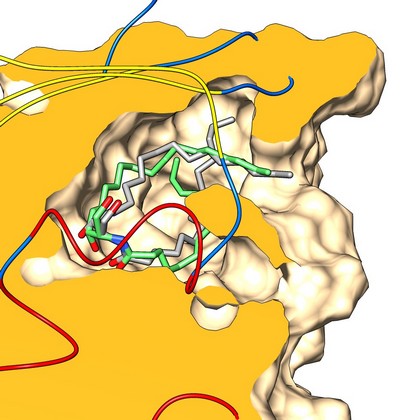
Stereospecific docking of long-chain amphiphilic derivatives
Superposition of the corresponding crystallographic structures (gray) with lowest energy docked poses (green) of C16-d-erythro-ceramide. The orientation and protein surface (orange) clipping show the placement of the lipid polar heads at the left of the binding site cavity of CERT (CERamide Transfer protein).
Superposition of the corresponding crystallographic structures (gray) with lowest energy docked poses (green) of C16-d-erythro-ceramide. The orientation and protein surface (orange) clipping show the placement of the lipid polar heads at the left of the binding site cavity of CERT (CERamide Transfer protein).
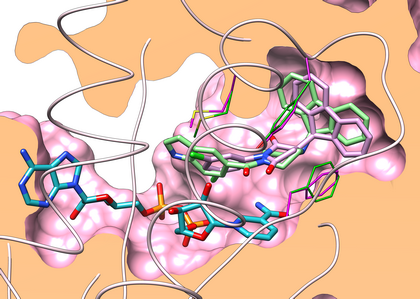
Poses (marine alcaloïds) resulting from docking with a Checkpoint Kinase
Clipped (UCSF Chimera) view of ATP-binding pocket of Chk1 (structure 1NVSa) including SB-218078 ligand (sand, mesh surface) and best poses of compounds 4 (light blue), 5 (purple) and 2 (light pink).
Clipped (UCSF Chimera) view of ATP-binding pocket of Chk1 (structure 1NVSa) including SB-218078 ligand (sand, mesh surface) and best poses of compounds 4 (light blue), 5 (purple) and 2 (light pink).
Succinimid based GEQ derivative
Superimposition of crystallographic conformation of GEQ (carbons in green stick representation) and simulated by docking binding mode of 11b (carbons in violet stick representation) in the binding groove of InhA (1P44 PDB entry, chain A). Degree of residue flexibility (side chains of residues around the site, backbone) was applied in docking procedure. Residues Phe149, Tyr158 and Met161 colored in magenta (docked protein structure) and green (1P44 protein structure).
Superimposition of crystallographic conformation of GEQ (carbons in green stick representation) and simulated by docking binding mode of 11b (carbons in violet stick representation) in the binding groove of InhA (1P44 PDB entry, chain A). Degree of residue flexibility (side chains of residues around the site, backbone) was applied in docking procedure. Residues Phe149, Tyr158 and Met161 colored in magenta (docked protein structure) and green (1P44 protein structure).
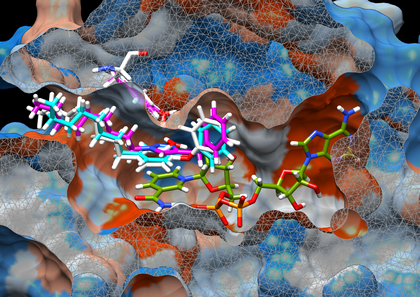
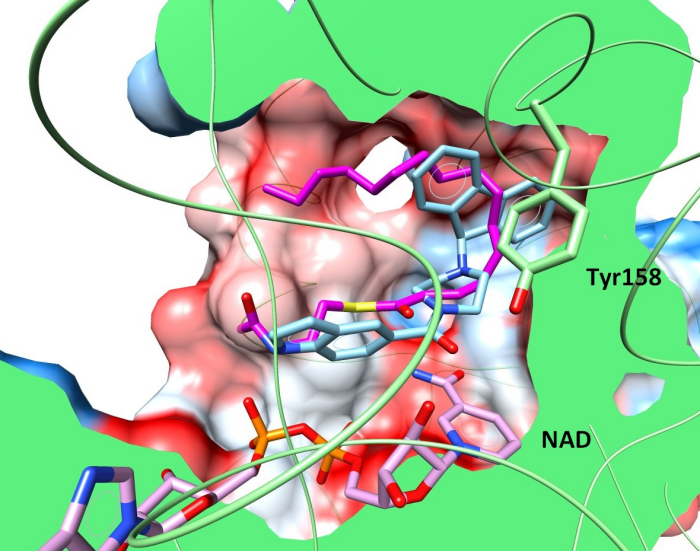
Triclosan derivatives docked in InhA minor portal
Hypothetical binding mode of compounds 8PS (cyan) and 18 (magenta) in the active site of InhA enzyme (PDB ID 2B37) obtained by molecular docking. Cocrystallised cofactor NAD+ and 8PS are coloured in green and white respectively; the molecular surface of protein is coloured using a hydrophobic scale and rendered using UCSF Chimera. Tyr158 residue is coloured (white for 2B37 native state) in a final conformation obtained after docking. The same calculation protocol was used in both cases and the docking of 8PS shows an agreement in reproducing crystallography (RMSD = 0.94 Å) without displacing Tyr158.
Hypothetical binding mode of compounds 8PS (cyan) and 18 (magenta) in the active site of InhA enzyme (PDB ID 2B37) obtained by molecular docking. Cocrystallised cofactor NAD+ and 8PS are coloured in green and white respectively; the molecular surface of protein is coloured using a hydrophobic scale and rendered using UCSF Chimera. Tyr158 residue is coloured (white for 2B37 native state) in a final conformation obtained after docking. The same calculation protocol was used in both cases and the docking of 8PS shows an agreement in reproducing crystallography (RMSD = 0.94 Å) without displacing Tyr158.
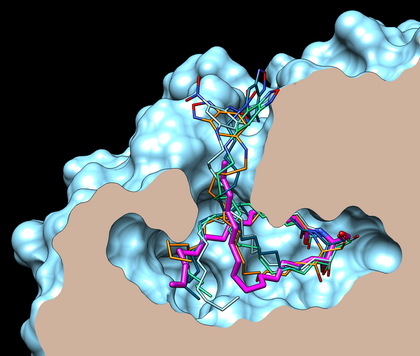
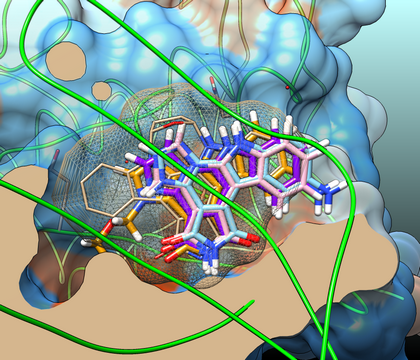
Binding modes of lipidic fluorescent probes
Putative conformations of probe A (n = 14) in which the NBD group is found outside CERT cavity (2E3N, blue). These docking poses (extracted from the 50 higher Molegro’s rerank scores) are typical of conformations that reproduce the conformation of hydroxyl and amido groups of a ceramide (right part of the site) with a good accuracy. The C16-ceramide (pink) crystallographic conformation (2E3O) is shown for reference.
Putative conformations of probe A (n = 14) in which the NBD group is found outside CERT cavity (2E3N, blue). These docking poses (extracted from the 50 higher Molegro’s rerank scores) are typical of conformations that reproduce the conformation of hydroxyl and amido groups of a ceramide (right part of the site) with a good accuracy. The C16-ceramide (pink) crystallographic conformation (2E3O) is shown for reference.
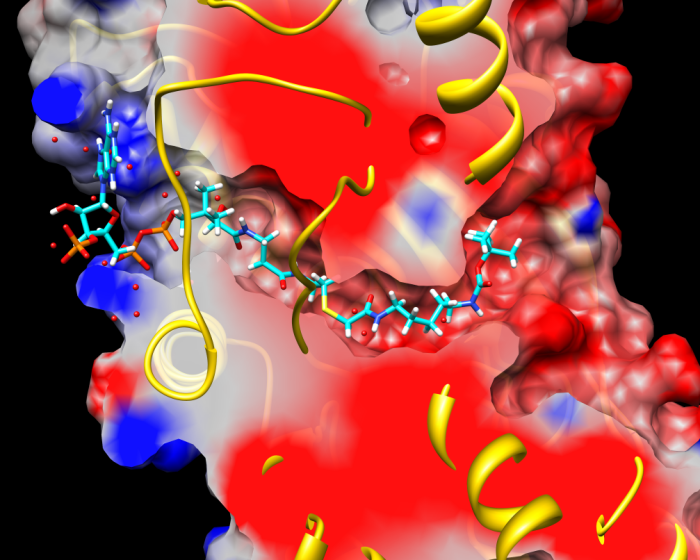
Bisubstrate inhibitor docked in histone acetyltransferase (p300)
Binding mode of best compound. Model clipping of PDB structure 3BIY with continuum electrostatics coloring of molecular surface, and compound 3 after docking (35,36). The figure shows occupancy of cavities P1 and P2 by long chain (n = 5) and Boc moieties, respectively, at right of the (yellow) overlapping loop (upper clipping plane).
Binding mode of best compound. Model clipping of PDB structure 3BIY with continuum electrostatics coloring of molecular surface, and compound 3 after docking (35,36). The figure shows occupancy of cavities P1 and P2 by long chain (n = 5) and Boc moieties, respectively, at right of the (yellow) overlapping loop (upper clipping plane).
Related references
- Hemisynthesis, Antitumoral Effect, and Molecular Docking Studies of Ferutinin and Its Analogues. Safi R, Rodriguez F, Hilal G, Diab-Assaf M, Diab Y, El-Sabban M, Najjar F, Delfourne E. Chem Biol Drug Des. (2016) 87(3):382-97.
- Design, synthesis and evaluation of new GEQ derivatives as inhibitors of InhA enzyme and Mycobacterium tuberculosis growth. Chollet A, Mori G, Menendez C, Rodriguez F, Fabing I, Pasca MR, Madacki J, Korduláková J, Constant P, Quémard A, Bernardes-Génisson V, Lherbet C, Baltas M. Eur J Med Chem. (2015) 28;101:218-35.
- Identification of Novel CERT Ligands as Potential Ceramide Trafficking Inhibitors. Santos C, Rodriguez F, Garcia V, Moravčikova D, Berkeš D, Daïch A, Levade T, Baudoin-Dehoux C, Ballereau S, Génisson Y. ChemBioChem (2014) 15: 2522 – 2528.
- Design of granulatimide and isogranulatimide analogues as potential Chk1 inhibitors: Study of amino-platforms for their synthesis. Lavrard H, Rodriguez F, Delfourne D. Bioorg. Med. Chem. (2014) 22:4961–4967.
- A biologically relevant ceramide fluorescent probe to assess the binding of potential ligands to the CERT transfer protein. Combemale S, Santos C, Rodriguez F, Garcia V, Galaup C, Frongia C, Lobjois V, Levade T, Baudoin-Dehoux C, Ballereau S, Génisson Y. RSC Adv. (2013) 3, 18970–18984.
- Design, chemical synthesis of 3-(9H-fluoren-9-yl)pyrrolidine-2,5-dione derivatives and biological activity against enoyl-ACP reductase (InhA) and Mycobacterium tuberculosis. Matviiuk T, Rodriguez F, Saffon N, Mallet-Ladeira S, Gorichko M, Jesus Lopes Ribeiro AL, Pasca MR, Lherbet C, Voitenko Z, Baltas M. Eur. J. Med. Chem. (2013) 70, 37-48.
- Chemical synthesis and biological evaluation of triazole derivatives as inhibitors of InhA and antituberculosis agents. Menendez C, Chollet A, Rodriguez F, Inard C, Pasca MR, Lherbet C, Baltas M. Eur. J. Med. Chem. (2012) 52, 275-283.
- New potent bisubstrate inhibitors of histone acetyltransferase p300: design, synthesis and biological evaluation. Kwie FH, Briet M, Soupaya D, Hoffmann P, Maturano M, Rodriguez F, Blonski C, Lherbet C, Baudoin-Dehoux C. Chem Biol Drug Des. (2011) 77(1):86-92.


